Description
In this course, you will :
- Describe the key technical features of data models using Entity-Relationship Diagrams (ERDs).
- We investigate the technical features of clinical data models using MIMIC3 as an example, and we investigate common data models using OMOP as an example.
- This module teaches students about the processes and challenges of extracting, transforming, and loading (ETL) data using real-world data and terminology mapping examples.
- We investigate the dimensions of data quality by reviewing its challenges, data quality measurements used to measure it, and data quality rules used to determine its usability.
Syllabus :
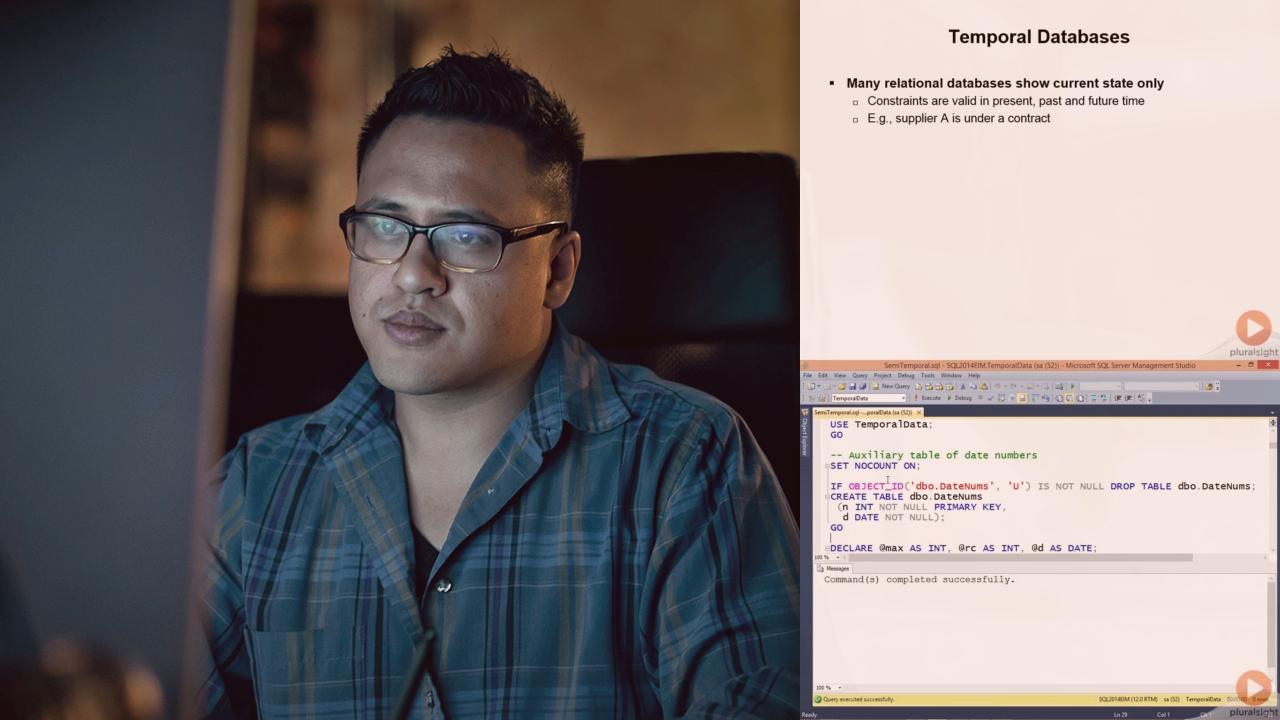
1. Introduction: Clinical Data Models and Common Data Models
- Clinical Research Data Warehouses
- Entity Relationship Diagrams (ERDs)
- Clinical Data Models
- Why Common Data Models?
- A Quick Tour of a Common Data Model: i2b
- A Quick Tour of a Common Data Model: OMOP
- A Quick Tour of a Common Data Model: Sentinel
- A Quick Tour of a Common Data Model: PCORNet
2. Tools: Querying Clinical Data Models
- A Deep Dive into the MIMIC-III Data Model
- Querying MIMIC-III
- A Deep Dive into OMOP Data Model
- Querying OMOP
- Comparing the MIMIC and OMOP Data Models
- The OHDSI Community Ecosystem
3. Techniques: Extract-Transform-Load and Terminology Mapping
- The ETL Task
- Structural versus Terminology Mapping
- Data Profiling with White Rabbit
- Data Mapping with the Rabbit in a Hat Tool
- Terminology Mapping
- Example mapping of MIMIC Patient to OMOP Person
4. Techniques: Data Quality Assessments
- Data quality dimensions / fitness for use
- Data profiling for data quality assessment
- Data quality assessment using SQL
- Callahan and Khare rules
- OHDSI Achilles and Achilles Heel
5. Practical Application: Create an ETL Process to Transform a MIMIC-III Table to OMOP
- Review of the ETL process
- Example: Transforming MIMIC Patient to OMOP Person